Faster Polynomial Features#
Links: notebook, html, PDF, python, slides, GitHub
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%matplotlib inline
Polynomial Features#
The current implementation of
PolynomialFeatures
(0.20.2) implements a term by term product for each pair
of features where
which is not
the most efficient way to do it.
import numpy.random
X = numpy.random.random((100, 5))
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(degree=2)
Xpoly = poly.fit_transform(X)
poly.get_feature_names()
['1',
'x0',
'x1',
'x2',
'x3',
'x4',
'x0^2',
'x0 x1',
'x0 x2',
'x0 x3',
'x0 x4',
'x1^2',
'x1 x2',
'x1 x3',
'x1 x4',
'x2^2',
'x2 x3',
'x2 x4',
'x3^2',
'x3 x4',
'x4^2']
%timeit poly.transform(X)
114 µs ± 12.4 µs per loop (mean ± std. dev. of 7 runs, 10000 loops each)
The class ExtendedFeatures implements a different way to compute the polynomial features as it tries to reduce the number of calls to numpy by using broacasted vector multplications.
from mlinsights.mlmodel import ExtendedFeatures
ext = ExtendedFeatures(poly_degree=2)
Xpoly = ext.fit_transform(X)
ext.get_feature_names()
['1',
'x0',
'x1',
'x2',
'x3',
'x4',
'x0^2',
'x0 x1',
'x0 x2',
'x0 x3',
'x0 x4',
'x1^2',
'x1 x2',
'x1 x3',
'x1 x4',
'x2^2',
'x2 x3',
'x2 x4',
'x3^2',
'x3 x4',
'x4^2']
%timeit ext.transform(X)
68.7 µs ± 10.6 µs per loop (mean ± std. dev. of 7 runs, 10000 loops each)
Comparison with 5 features#
from cpyquickhelper.numbers import measure_time
res = []
for n in [1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000,
5000, 10000, 20000, 50000, 100000, 200000]:
X = numpy.random.random((n, 5))
poly.fit(X)
ext.fit(X)
r1 = measure_time("poly.transform(X)", context=dict(X=X, poly=poly), repeat=5, number=10, div_by_number=True)
r2 = measure_time("ext.transform(X)", context=dict(X=X, ext=ext), repeat=5, number=10, div_by_number=True)
r3 = measure_time("poly.fit_transform(X)", context=dict(X=X, poly=poly), repeat=5, number=10, div_by_number=True)
r4 = measure_time("ext.fit_transform(X)", context=dict(X=X, ext=ext), repeat=5, number=10, div_by_number=True)
r1["name"] = "poly"
r2["name"] = "ext"
r3["name"] = "poly+fit"
r4["name"] = "ext+fit"
r1["size"] = n
r2["size"] = n
r3["size"] = n
r4["size"] = n
res.append(r1)
res.append(r2)
res.append(r3)
res.append(r4)
import pandas
df = pandas.DataFrame(res)
df.tail()
| average | deviation | min_exec | max_exec | repeat | number | context_size | name | size | |
|---|---|---|---|---|---|---|---|---|---|
| 63 | 0.037830 | 0.005577 | 0.031248 | 0.044832 | 5 | 10 | 240 | ext+fit | 100000 |
| 64 | 0.072671 | 0.005360 | 0.067559 | 0.082539 | 5 | 10 | 240 | poly | 200000 |
| 65 | 0.075712 | 0.018271 | 0.060476 | 0.100143 | 5 | 10 | 240 | ext | 200000 |
| 66 | 0.106755 | 0.019861 | 0.079880 | 0.139184 | 5 | 10 | 240 | poly+fit | 200000 |
| 67 | 0.074090 | 0.009142 | 0.063925 | 0.085899 | 5 | 10 | 240 | ext+fit | 200000 |
piv = df.pivot("size", "name", "average")
piv[:5]
| name | ext | ext+fit | poly | poly+fit |
|---|---|---|---|---|
| size | ||||
| 1 | 0.000068 | 0.000402 | 0.000238 | 0.000275 |
| 2 | 0.000066 | 0.000156 | 0.000166 | 0.000213 |
| 5 | 0.000031 | 0.000427 | 0.000165 | 0.000196 |
| 10 | 0.000048 | 0.000237 | 0.000134 | 0.000306 |
| 20 | 0.000070 | 0.000188 | 0.000109 | 0.000153 |
ax = piv.plot(logy=True, logx=True)
ax.set_title("Polynomial Features for 5 features\ndegree=2")
ax.set_ylabel("seconds")
ax.set_xlabel("number of observations");
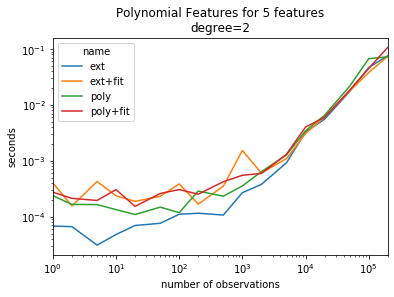
The gain is mostly visible for small dimensions.
Comparison with 1000 observations#
In this experiment, the number of observations is fixed to 1000 but the number of features varies.
poly = PolynomialFeatures(degree=2)
ext = ExtendedFeatures(poly_degree=2)
# implementation of PolynomialFeatures in 0.20.2
extslow = ExtendedFeatures(poly_degree=2, kind="poly-slow")
res = []
for n in [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 40, 50]:
X = numpy.random.random((1000, n))
poly.fit(X)
ext.fit(X)
extslow.fit(X)
r1 = measure_time("poly.transform(X)", context=dict(X=X, poly=poly), repeat=5, number=30, div_by_number=True)
r2 = measure_time("ext.transform(X)", context=dict(X=X, ext=ext), repeat=5, number=30, div_by_number=True)
r3 = measure_time("extslow.transform(X)", context=dict(X=X, extslow=extslow), repeat=5, number=30, div_by_number=True)
r1["name"] = "poly"
r2["name"] = "ext"
r3["name"] = "extslow"
r1["nfeat"] = n
r2["nfeat"] = n
r3["nfeat"] = n
x1 = poly.transform(X)
x2 = ext.transform(X)
x3 = extslow.transform(X)
r1["numf"] = x1.shape[1]
r2["numf"] = x2.shape[1]
r3["numf"] = x3.shape[1]
res.append(r1)
res.append(r2)
res.append(r3)
import pandas
df = pandas.DataFrame(res)
df.tail()
| average | deviation | min_exec | max_exec | repeat | number | context_size | name | nfeat | numf | |
|---|---|---|---|---|---|---|---|---|---|---|
| 37 | 0.009331 | 0.001603 | 0.008280 | 0.012519 | 5 | 30 | 240 | ext | 40 | 861 |
| 38 | 0.022619 | 0.002868 | 0.018793 | 0.026324 | 5 | 30 | 240 | extslow | 40 | 861 |
| 39 | 0.013188 | 0.000370 | 0.012828 | 0.013888 | 5 | 30 | 240 | poly | 50 | 1326 |
| 40 | 0.012817 | 0.000102 | 0.012700 | 0.012951 | 5 | 30 | 240 | ext | 50 | 1326 |
| 41 | 0.030384 | 0.000717 | 0.029955 | 0.031813 | 5 | 30 | 240 | extslow | 50 | 1326 |
piv = df.pivot("nfeat", "name", "average")
piv[:5]
| name | ext | extslow | poly |
|---|---|---|---|
| nfeat | |||
| 1 | 0.000026 | 0.000059 | 0.000152 |
| 2 | 0.000055 | 0.000100 | 0.000113 |
| 3 | 0.000161 | 0.000381 | 0.000237 |
| 4 | 0.000148 | 0.000221 | 0.000219 |
| 5 | 0.000185 | 0.000340 | 0.000236 |
ax = piv.plot(logy=True, logx=True)
ax.set_title("Polynomial Features for 1000 observations\ndegree=2")
ax.set_ylabel("seconds")
ax.set_xlabel("number of features");
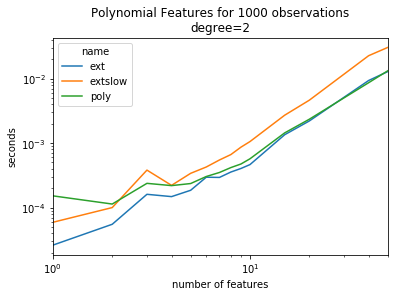
It is twice faster.
Comparison for different degrees#
In this experiment, the number of observations and features is fixed, the degree increases.
res = []
for n in [2, 3, 4, 5, 6, 7, 8]:
X = numpy.random.random((1000, 4))
poly = PolynomialFeatures(degree=n)
ext = ExtendedFeatures(poly_degree=n)
poly.fit(X)
ext.fit(X)
r1 = measure_time("poly.transform(X)", context=dict(X=X, poly=poly), repeat=5, number=30, div_by_number=True)
r2 = measure_time("ext.transform(X)", context=dict(X=X, ext=ext), repeat=5, number=30, div_by_number=True)
r1["name"] = "poly"
r2["name"] = "ext"
r1["degree"] = n
r2["degree"] = n
x1 = poly.transform(X)
x2 = ext.transform(X)
r1["numf"] = x1.shape[1]
r2["numf"] = x2.shape[1]
res.append(r1)
res.append(r2)
import pandas
df = pandas.DataFrame(res)
df.tail()
| average | deviation | min_exec | max_exec | repeat | number | context_size | name | degree | numf | |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 0.001960 | 0.000067 | 0.001915 | 0.002094 | 5 | 30 | 240 | ext | 6 | 210 |
| 10 | 0.003131 | 0.000118 | 0.003009 | 0.003327 | 5 | 30 | 240 | poly | 7 | 330 |
| 11 | 0.003076 | 0.000233 | 0.002845 | 0.003393 | 5 | 30 | 240 | ext | 7 | 330 |
| 12 | 0.004299 | 0.000046 | 0.004243 | 0.004367 | 5 | 30 | 240 | poly | 8 | 495 |
| 13 | 0.004157 | 0.000035 | 0.004114 | 0.004217 | 5 | 30 | 240 | ext | 8 | 495 |
piv = df.pivot("degree", "name", "average")
piv[:5]
| name | ext | poly |
|---|---|---|
| degree | ||
| 2 | 0.000140 | 0.000312 |
| 3 | 0.000304 | 0.000363 |
| 4 | 0.000506 | 0.000579 |
| 5 | 0.000715 | 0.000789 |
| 6 | 0.001960 | 0.002032 |
ax = piv.plot(logy=True, logx=True)
ax.set_title("Polynomial Features for 1000 observations\nnumber of features is 4")
ax.set_ylabel("seconds")
ax.set_xlabel("degree");
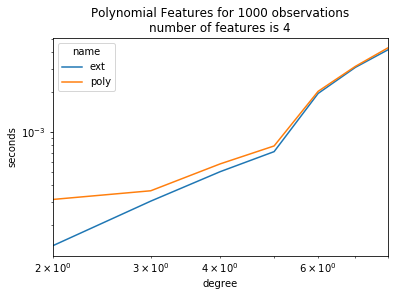
It is worth transposing.
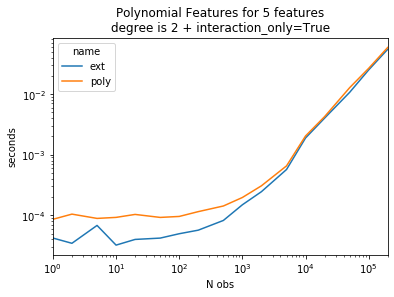
Same experiment with interaction_only=True#
res = []
for n in [1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000,
5000, 10000, 20000, 50000, 100000, 200000]:
poly = PolynomialFeatures(degree=2, interaction_only=True)
ext = ExtendedFeatures(poly_degree=2, poly_interaction_only=True)
X = numpy.random.random((n, 5))
poly.fit(X)
ext.fit(X)
r1 = measure_time("poly.transform(X)", context=dict(X=X, poly=poly), repeat=2, number=30, div_by_number=True)
r2 = measure_time("ext.transform(X)", context=dict(X=X, ext=ext), repeat=2, number=30, div_by_number=True)
r1["name"] = "poly"
r2["name"] = "ext"
r1["size"] = n
r2["size"] = n
res.append(r1)
res.append(r2)
import pandas
df = pandas.DataFrame(res)
df.tail()
| average | deviation | min_exec | max_exec | repeat | number | context_size | name | size | |
|---|---|---|---|---|---|---|---|---|---|
| 29 | 0.010691 | 0.000073 | 0.010618 | 0.010764 | 2 | 30 | 240 | ext | 50000 |
| 30 | 0.026612 | 0.000794 | 0.025817 | 0.027406 | 2 | 30 | 240 | poly | 100000 |
| 31 | 0.025052 | 0.001583 | 0.023469 | 0.026635 | 2 | 30 | 240 | ext | 100000 |
| 32 | 0.058772 | 0.001345 | 0.057427 | 0.060118 | 2 | 30 | 240 | poly | 200000 |
| 33 | 0.054771 | 0.004555 | 0.050216 | 0.059327 | 2 | 30 | 240 | ext | 200000 |
piv = df.pivot("size", "name", "average")
piv[:5]
| name | ext | poly |
|---|---|---|
| size | ||
| 1 | 0.000042 | 0.000086 |
| 2 | 0.000034 | 0.000104 |
| 5 | 0.000068 | 0.000089 |
| 10 | 0.000032 | 0.000092 |
| 20 | 0.000040 | 0.000103 |
ax = piv.plot(logy=True, logx=True)
ax.set_title("Polynomial Features for 5 features\ndegree is 2 + interaction_only=True")
ax.set_ylabel("seconds")
ax.set_xlabel("N obs");
Memory profiler#
from memory_profiler import memory_usage
poly = PolynomialFeatures(degree=2, interaction_only=True)
poly.fit(X)
memory_usage((poly.transform, (X,)), interval=0.1, max_usage=True)
258.02734375
def pick_value(v):
try:
return v[0]
except TypeError:
return v
res = []
for n in [10000, 50000, 100000, 200000]:
X = numpy.random.random((n, 50))
print(n)
poly = PolynomialFeatures(degree=2, interaction_only=True)
ext = ExtendedFeatures(poly_degree=2, poly_interaction_only=True)
poly.fit(X)
ext.fit(X)
r1 = memory_usage((poly.transform, (X,)), interval=0.1, max_usage=True)
r2 = memory_usage((ext.transform, (X,)), interval=0.1, max_usage=True)
r1 = {"memory": pick_value(r1)}
r2 = {"memory": pick_value(r2)}
r1["name"] = "poly"
r2["name"] = "ext"
r1["size"] = n
r2["size"] = n
res.append(r1)
res.append(r2)
import pandas
df = pandas.DataFrame(res)
df.tail()
10000
50000
100000
200000
| memory | name | size | |
|---|---|---|---|
| 3 | 699.679688 | ext | 50000 |
| 4 | 1243.664062 | poly | 100000 |
| 5 | 1205.515625 | ext | 100000 |
| 6 | 1952.316406 | poly | 200000 |
| 7 | 2029.765625 | ext | 200000 |
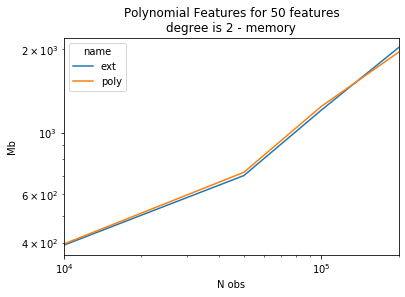
piv = df.pivot("size", "name", "memory")
piv[:5]
| name | ext | poly |
|---|---|---|
| size | ||
| 10000 | 392.445312 | 396.347656 |
| 50000 | 699.679688 | 718.839844 |
| 100000 | 1205.515625 | 1243.664062 |
| 200000 | 2029.765625 | 1952.316406 |
ax = piv.plot(logy=True, logx=True)
ax.set_title("Polynomial Features for 50 features\ndegree is 2 - memory")
ax.set_ylabel("Mb")
ax.set_xlabel("N obs");