Un arbre de décision en réseaux de neurones#
Links: notebook, html, PDF, python, slides, GitHub
L’idée est de convertir sous la forme d’un réseaux de neurones un arbre de décision puis de continuer l’apprentissage de façon à obtenir un assemblage de régression logistique plutôt que de décision binaire.
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%matplotlib inline
from jyquickhelper import RenderJsDot
import numpy
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from tqdm import tqdm
Un exemple sur Iris#
La méthode ne marche que sur un problème de classification binaire.
from sklearn.datasets import load_iris
data = load_iris()
X, y = data.data[:, :2], data.target
y = y % 2
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=11)
from sklearn.tree import DecisionTreeClassifier
dec = DecisionTreeClassifier(max_depth=2, random_state=11)
dec.fit(X_train, y_train)
dec.score(X_test, y_test)
0.6052631578947368
from sklearn.tree import export_graphviz
dot = export_graphviz(dec, filled=True)
dot = dot.replace("shape=box, ", "shape=box, fontsize=10, ")
RenderJsDot(dot)
print(dot)
digraph Tree {
node [shape=box, fontsize=10, style="filled", color="black", fontname="helvetica"] ;
edge [fontname="helvetica"] ;
0 [label="x[1] <= 2.95ngini = 0.454nsamples = 112nvalue = [73, 39]", fillcolor="#f3c4a3"] ;
1 [label="x[0] <= 7.05ngini = 0.429nsamples = 45nvalue = [14, 31]", fillcolor="#92c9f1"] ;
0 -> 1 [labeldistance=2.5, labelangle=45, headlabel="True"] ;
2 [label="gini = 0.402nsamples = 43nvalue = [12, 31]", fillcolor="#86c3ef"] ;
1 -> 2 ;
3 [label="gini = 0.0nsamples = 2nvalue = [2, 0]", fillcolor="#e58139"] ;
1 -> 3 ;
4 [label="x[1] <= 3.25ngini = 0.21nsamples = 67nvalue = [59, 8]", fillcolor="#e99254"] ;
0 -> 4 [labeldistance=2.5, labelangle=-45, headlabel="False"] ;
5 [label="gini = 0.375nsamples = 32nvalue = [24, 8]", fillcolor="#eeab7b"] ;
4 -> 5 ;
6 [label="gini = 0.0nsamples = 35nvalue = [35, 0]", fillcolor="#e58139"] ;
4 -> 6 ;
}
L’arbre de décision est petit donc visuellement réduit et il est perfectible aussi.
Même exemple en réseau de neurones#
Chaque noeud de l’arbre de décision est converti en deux neurones : *
un qui le relie à l’entrée et qui évalue la décision, il produit la
valeur * un autre qui associe le résultat du premier noeud
avec celui le précède dans la structure de l’arbre de décision, il
produit la valeur
La décision finale est quelque chose comme
. Un neurone agrège le résultat de toutes
les feuilles.
from mlstatpy.ml.neural_tree import NeuralTreeNet
net = NeuralTreeNet.create_from_tree(dec)
RenderJsDot(net.to_dot())
On considère une entrée en particulier.
n = 60
dec.predict_proba(X[n: n+1])
array([[0.27906977, 0.72093023]])
Les sorties du réseau de neurones :
net.predict(X[n: n+1])[:, -2:]
array([[0.12536069, 0.87463931]])
Et on trace les valeurs intermédiaires.
RenderJsDot(net.to_dot(X=X[n]))
On poursuit la comparaison :
dec.predict_proba(X_test)[:5]
array([[0.75 , 0.25 ],
[0.75 , 0.25 ],
[0.27906977, 0.72093023],
[1. , 0. ],
[0.27906977, 0.72093023]])
net.predict(X_test)[:5, -2:]
array([[0.79156817, 0.20843183],
[0.73646978, 0.26353022],
[0.29946111, 0.70053889],
[0.94070094, 0.05929906],
[0.24924737, 0.75075263]])
dec.predict_proba(X_test)[-5:]
array([[1. , 0. ],
[0.75, 0.25],
[1. , 0. ],
[0.75, 0.25],
[0.75, 0.25]])
net.predict(X_test)[-5:, -2:]
array([[0.93247891, 0.06752109],
[0.86338585, 0.13661415],
[0.98219036, 0.01780964],
[0.98352807, 0.01647193],
[0.73646978, 0.26353022]])
numpy.argmax(net.predict(X_test)[-5:, -2:], axis=1)
array([0, 0, 0, 0, 0], dtype=int64)
On compare visuellement les deux frontières de classification.
def plot_grid(X, y, fct, title, ax=None):
cmap_light = ListedColormap(['orange', 'cyan', 'cornflowerblue'])
cmap_bold = ListedColormap(['darkorange', 'c', 'darkblue'])
h = .05
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = numpy.meshgrid(numpy.arange(x_min, x_max, h),
numpy.arange(y_min, y_max, h))
Z = fct(numpy.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
if ax is None:
_, ax = plt.subplots(1, 1)
ax.pcolormesh(xx, yy, Z, cmap=cmap_light)
ax.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap_bold,
edgecolor='k', s=20)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_title(title)
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_grid(X, y, dec.predict, dec.__class__.__name__, ax=ax[0])
plot_grid(X, y,
lambda x: numpy.argmax(net.predict(x)[:, -2:], axis=1),
net.__class__.__name__, ax=ax[1])
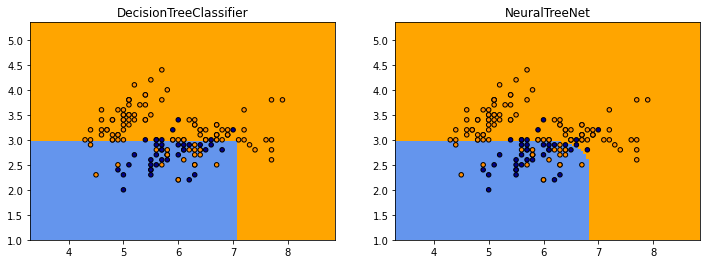
Le code qui produit les prédictions du réseau de neurones est assez long à exécuter mais il produit à peu près les mêmes frontières excepté qu’elles sont plus arrondies.
Intermède de simples neurones de régression#
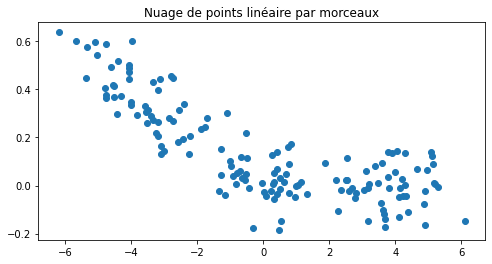
Avant d’apprendre ou plutôt de continuer l’apprentissage des coefficients du réseaux de neurones, voyons comment un neurone se débrouille sur un problème de régression. Le neurone n’est pas converti, il est appris.
regX = numpy.empty((150, 1), dtype=numpy.float64)
regX[:50, 0] = numpy.random.randn(50) - 4
regX[50:100, 0] = numpy.random.randn(50)
regX[100:, 0] = numpy.random.randn(50) + 4
noise = numpy.random.randn(regX.shape[0]) / 10
regY = regX[:, 0] * -0.5 * 0.2 + noise
regY[regX[:, 0] > 0.3] = noise[regX[:, 0] > 0.3]
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.scatter(regX[:, 0], regY)
ax.set_title("Nuage de points linéaire par morceaux");
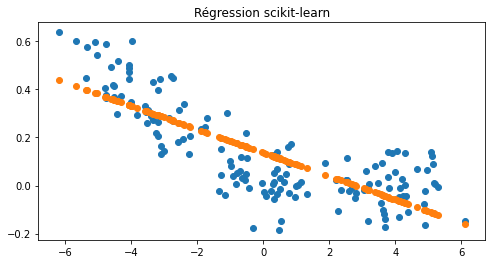
On cale une régression avec scikit-learn.
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(regX, regY)
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.scatter(regX[:, 0], regY)
ax.scatter(regX[:, 0], lr.predict(regX))
ax.set_title("Régression scikit-learn");
Et maintenant un neurone avec une fonction d’activation “identity”.
from mlstatpy.ml.neural_tree import NeuralTreeNode
neu = NeuralTreeNode(1, activation="identity")
neu
NeuralTreeNode(weights=array([0.98477397]), bias=0.8682318535624896, activation='identity')
neu.fit(regX, regY, verbose=True, max_iter=20)
0/20: loss: 1777 lr=0.002 max(coef): 0.98 l1=0/1.9 l2=0/1.7
1/20: loss: 2.705 lr=0.000163 max(coef): 0.18 l1=0.74/0.24 l2=0.33/0.035
2/20: loss: 2.223 lr=0.000115 max(coef): 0.16 l1=0.048/0.21 l2=0.0016/0.027
3/20: loss: 2.206 lr=9.42e-05 max(coef): 0.15 l1=0.49/0.2 l2=0.13/0.026
4/20: loss: 2.176 lr=8.16e-05 max(coef): 0.15 l1=0.81/0.2 l2=0.47/0.024
5/20: loss: 2.167 lr=7.3e-05 max(coef): 0.14 l1=0.19/0.19 l2=0.018/0.023
6/20: loss: 2.186 lr=6.66e-05 max(coef): 0.14 l1=2.1/0.2 l2=3.1/0.024
7/20: loss: 2.183 lr=6.17e-05 max(coef): 0.14 l1=1.4/0.2 l2=1.3/0.023
8/20: loss: 2.159 lr=5.77e-05 max(coef): 0.14 l1=1.2/0.19 l2=0.97/0.022
9/20: loss: 2.164 lr=5.44e-05 max(coef): 0.14 l1=0.53/0.19 l2=0.17/0.022
10/20: loss: 2.173 lr=5.16e-05 max(coef): 0.14 l1=1.1/0.19 l2=0.77/0.023
11/20: loss: 2.16 lr=4.92e-05 max(coef): 0.14 l1=0.61/0.19 l2=0.22/0.022
12/20: loss: 2.162 lr=4.71e-05 max(coef): 0.14 l1=1/0.19 l2=0.55/0.022
13/20: loss: 2.161 lr=4.53e-05 max(coef): 0.14 l1=0.86/0.19 l2=0.43/0.022
14/20: loss: 2.173 lr=4.36e-05 max(coef): 0.14 l1=0.12/0.19 l2=0.0077/0.022
15/20: loss: 2.158 lr=4.22e-05 max(coef): 0.14 l1=0.049/0.19 l2=0.0017/0.022
16/20: loss: 2.157 lr=4.08e-05 max(coef): 0.14 l1=0.14/0.19 l2=0.011/0.022
17/20: loss: 2.169 lr=3.96e-05 max(coef): 0.14 l1=0.46/0.19 l2=0.11/0.022
18/20: loss: 2.17 lr=3.85e-05 max(coef): 0.14 l1=0.52/0.18 l2=0.16/0.021
19/20: loss: 2.157 lr=3.75e-05 max(coef): 0.14 l1=0.1/0.19 l2=0.0055/0.022
20/20: loss: 2.166 lr=3.65e-05 max(coef): 0.14 l1=0.75/0.19 l2=0.33/0.022
NeuralTreeNode(weights=array([-0.04865864]), bias=0.138494368212747, activation='identity')
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.scatter(regX[:, 0], regY)
ax.scatter(regX[:, 0], lr.predict(regX), label="sklearn")
ax.scatter(regX[:, 0], neu.predict(regX), label="NeuralTreeNode")
ax.legend()
ax.set_title("Régression et neurones");
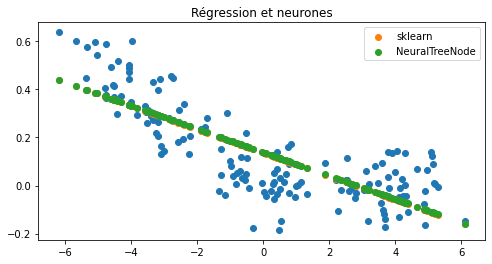
Ca marche. Et avec d’autres fonctions d’activation…
neus = {'identity': neu}
for act in tqdm(['relu', 'leakyrelu', 'sigmoid']):
nact = NeuralTreeNode(1, activation=act)
nact.fit(regX, regY)
neus[act] = nact
100%|██████████| 3/3 [00:01<00:00, 1.78it/s]
neus['relu'], neus['leakyrelu']
(NeuralTreeNode(weights=array([-0.22418012]), bias=-0.5116596757581577, activation='relu'),
NeuralTreeNode(weights=array([-0.01985412]), bias=-0.3531607623488657, activation='leakyrelu'))
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.scatter(regX[:, 0], regY)
ax.scatter(regX[:, 0], lr.predict(regX), label="sklearn")
for k, v in neus.items():
ax.scatter(regX[:, 0], v.predict(regX), label=k)
ax.legend()
ax.set_title("Régression, neurone\nactivation");
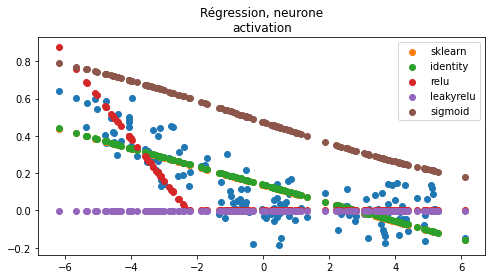
Rien de surprenant. La fonction sigmoïde prend ses valeurs entre 0 et 1. La fonction relu est parfois nulle sur une demi-droite, dès que la fonction est nulle sur l’ensemble du nuage de points, le gradient est nul partout (voir Rectifier (neural networks)). La fonction leaky relu est définie comme suit :
Le gradient n’est pas nul sur la partie la plus plate.
Intermède de simples neurones de classification#
Avant d’apprendre ou plutôt de continuer l’apprentissage des coefficients du réseaux de neurones, voyons comment un neurone se débrouille sur un problème de classification. Le neurone n’est pas converti mais appris.
from sklearn.linear_model import LogisticRegression
clsX = numpy.empty((100, 2), dtype=numpy.float64)
clsX[:50] = numpy.random.randn(50, 2)
clsX[50:] = numpy.random.randn(50, 2) + 2
clsy = numpy.zeros(100, dtype=numpy.int64)
clsy[50:] = 1
logr = LogisticRegression()
logr.fit(clsX, clsy)
pred1 = logr.predict(clsX)
def line_cls(x0, x1, coef, bias):
y0 = -(coef[0,0] * x0 + bias) / coef[0,1]
y1 = -(coef[0,0] * x1 + bias) / coef[0,1]
return x0, y0, x1, y1
x0, y0, x1, y1 = line_cls(-5, 5, logr.coef_, logr.intercept_)
h = 0.1
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.scatter(clsX[clsy == 0, 0], clsX[clsy == 0, 1], label='cl0')
ax.scatter(clsX[clsy == 1, 0], clsX[clsy == 1, 1], label='cl1')
ax.scatter(clsX[pred1 == 0, 0] + h, clsX[pred1 == 0, 1] + h, label='LR0')
ax.scatter(clsX[pred1 == 1, 0] + h, clsX[pred1 == 1, 1] + h, label='LR1')
ax.plot([x0, x1], [y0, y1], 'y--', lw=4, label='frontière LR')
ax.set_ylim([-3, 3])
ax.legend()
ax.set_title("Classification et neurones");
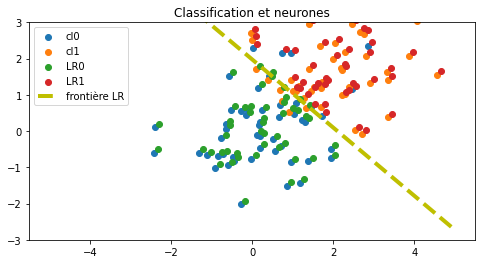
Un neurone de classification binaire produit deux sorties, une pour chaque classe, et sont normalisées à 1. La fonction d’activation est la fonction softmax.
clsY = numpy.empty((clsy.shape[0], 2), dtype=numpy.float64)
clsY[:, 1] = clsy
clsY[:, 0] = 1 - clsy
softneu = NeuralTreeNode(2, activation='softmax')
softneu
NeuralTreeNode(weights=array([[ 1.88671701, -1.05183379],
[-1.10109824, 0.71717564]]), bias=array([ 0.57621979, -1.30925916]), activation='softmax')
softneu.fit(clsX, clsY, verbose=True, max_iter=20, lr=0.001)
0/20: loss: 876.1 lr=0.001 max(coef): 1.9 l1=0/6.6 l2=0/8.4
1/20: loss: 274.4 lr=9.95e-05 max(coef): 4.3 l1=52/17 l2=6.1e+02/58
2/20: loss: 262.7 lr=7.05e-05 max(coef): 5.2 l1=21/21 l2=84/87
3/20: loss: 257.9 lr=5.76e-05 max(coef): 5.5 l1=20/22 l2=1.6e+02/99
4/20: loss: 255.3 lr=4.99e-05 max(coef): 5.8 l1=29/24 l2=1.5e+02/1.1e+02
5/20: loss: 253 lr=4.47e-05 max(coef): 6.1 l1=43/25 l2=3.8e+02/1.2e+02
6/20: loss: 251 lr=4.08e-05 max(coef): 6.3 l1=16/26 l2=82/1.3e+02
7/20: loss: 249.6 lr=3.78e-05 max(coef): 6.5 l1=31/27 l2=2.1e+02/1.4e+02
8/20: loss: 248.1 lr=3.53e-05 max(coef): 6.6 l1=48/28 l2=4.9e+02/1.4e+02
9/20: loss: 247.5 lr=3.33e-05 max(coef): 6.8 l1=32/28 l2=3.1e+02/1.5e+02
10/20: loss: 246.3 lr=3.16e-05 max(coef): 7 l1=31/29 l2=2e+02/1.6e+02
11/20: loss: 245.4 lr=3.01e-05 max(coef): 7.1 l1=15/30 l2=75/1.7e+02
12/20: loss: 243.4 lr=2.89e-05 max(coef): 7.3 l1=89/30 l2=1.5e+03/1.7e+02
13/20: loss: 242.1 lr=2.77e-05 max(coef): 7.4 l1=20/31 l2=94/1.8e+02
14/20: loss: 240.5 lr=2.67e-05 max(coef): 7.6 l1=22/31 l2=99/1.8e+02
15/20: loss: 239.7 lr=2.58e-05 max(coef): 7.7 l1=17/32 l2=74/1.9e+02
16/20: loss: 239.6 lr=2.5e-05 max(coef): 7.8 l1=37/33 l2=2.7e+02/2e+02
17/20: loss: 239.2 lr=2.42e-05 max(coef): 7.9 l1=43/33 l2=4.2e+02/2e+02
18/20: loss: 238.1 lr=2.36e-05 max(coef): 8 l1=46/34 l2=4.6e+02/2.1e+02
19/20: loss: 237.2 lr=2.29e-05 max(coef): 8.1 l1=20/34 l2=1.5e+02/2.2e+02
20/20: loss: 236.3 lr=2.24e-05 max(coef): 8.2 l1=39/35 l2=2.8e+02/2.2e+02
NeuralTreeNode(weights=array([[5.81047306, 2.25156457],
[7.51026475, 6.01918094]]), bias=array([8.23116261, 4.70093726]), activation='softmax')
pred = softneu.predict(clsX)
pred[:5]
array([[9.87624236e-01, 1.23757641e-02],
[9.99369242e-01, 6.30757836e-04],
[9.99641273e-01, 3.58726942e-04],
[9.55535770e-01, 4.44642301e-02],
[9.99991083e-01, 8.91676982e-06]])
pred2 = (pred[:, 1] > 0.5).astype(numpy.int64)
x00, y00, x01, y01 = line_cls(-4, 4, softneu.coef[:1, 1:], softneu.bias[0])
x10, y10, x11, y11 = line_cls(-4, 4, softneu.coef[1:, 1:], softneu.bias[1])
xa, ya, xb, yb = line_cls(
-5, 5, softneu.coef[1:, 1:] - softneu.coef[:1, 1:],
softneu.bias[1] - softneu.bias[0])
fig, ax = plt.subplots(1, 2, figsize=(14, 6))
for i in [0, 1]:
ax[i].scatter(clsX[clsy == 0, 0], clsX[clsy == 0, 1], label='cl0')
ax[i].scatter(clsX[clsy == 1, 0], clsX[clsy == 1, 1], label='cl1')
ax[i].scatter(clsX[pred1 == 0, 0] + h, clsX[pred1 == 0, 1] + h, label='LR0')
ax[i].scatter(clsX[pred1 == 1, 0] + h, clsX[pred1 == 1, 1] + h, label='LR1')
ax[i].scatter(clsX[pred2 == 0, 0] + h, clsX[pred2 == 0, 1] - h, label='NN0')
ax[i].scatter(clsX[pred2 == 1, 0] + h, clsX[pred2 == 1, 1] - h, label='NN1')
ax[0].plot([x0, x1], [y0, y1], 'y--', lw=4, label='frontière LR')
ax[1].plot([x00, x01], [y00, y01], 'r--', lw=4, label='droite neurone 0')
ax[1].plot([x10, x11], [y10, y11], 'b--', lw=4, label='droite neurone 1')
ax[0].plot([xa, xb], [ya, yb], 'c--', lw=4, label='frontière neurone')
ax[0].set_ylim([max(-6, min([-3, y10, y11, y11, y01])),
min(6, max([3, y10, y11, y11, y01]))])
ax[1].set_ylim([max(-6, min([-3, y10, y11, y11, y01])),
min(6, max([3, y10, y11, y11, y01]))])
ax[0].legend()
ax[1].legend()
ax[0].set_title("Frontière de classification")
ax[1].set_title("Neurones");
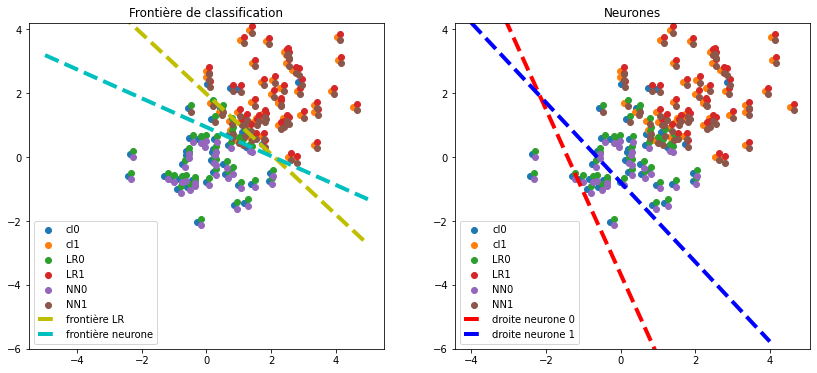
Ca marche. On vérifie en calculant le score. Le neurone a deux sorties. La frontière est définie par l’ensemble des points pour lesquels les deux sorties sont égales. Par conséquent, la distance entre les deux droites définies par les coefficients du neurone doivent être égales. Il existe une infinité de solutions menant à la même frontière. On pourrait pénaliser les coefficients pour converger toujours vers la même solution.
from sklearn.metrics import roc_auc_score
roc_auc_score(clsy, logr.predict_proba(clsX)[:, 1])
0.944
roc_auc_score(clsy, softneu.predict(clsX)[:, 1])
0.9328000000000001
La performance est quasiment identique. Que ce soit la régression ou la classification, l’apprentissage d’un neurone fonctionne. En sera-t-il de même pour un assemblage de neurones ?
Apprentissage du réseau de neurones#
Maintenant qu’on a vu les différentes fonctions d’activations et leur application sur des problèmes simples, on revient aux arbres convertis sous la forme d’un réseau de neurones. La prochaine étape est de pouvoir améliorer les performances du modèle issu de la conversion d’un arbre de classification avec un algorithme du gradient. On construit pour cela un nuage de points un peu traficoté.
clsX = numpy.empty((150, 2), dtype=numpy.float64)
clsX[:100] = numpy.random.randn(100, 2)
clsX[:20, 0] -= 1
clsX[20:40, 0] -= 0.8
clsX[:100, 1] /= 2
clsX[:100, 1] += clsX[:100, 0] ** 2
clsX[100:] = numpy.random.randn(50, 2)
clsX[100:, 0] /= 2
clsX[100:, 1] += 2.5
clsy = numpy.zeros(X.shape[0], dtype=numpy.int64)
clsy[100:] = 1
logr = LogisticRegression()
logr.fit(clsX, clsy)
pred1 = logr.predict(clsX)
logr.score(clsX, clsy)
0.7133333333333334
x0, y0, x1, y1 = line_cls(-3, 3, logr.coef_, logr.intercept_)
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
plot_grid(clsX, clsy, logr.predict, logr.__class__.__name__, ax=ax)
ax.plot([x0, x1], [y0, y1], 'y--', lw=4, label='frontière LR');
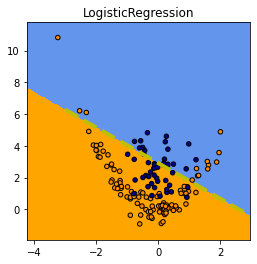
Même chose avec un arbre de décision et le réseau de neurones converti.
dec = DecisionTreeClassifier(max_depth=2)
dec.fit(clsX, clsy)
pred2 = dec.predict(clsX)
dec.score(clsX, clsy)
0.8733333333333333
On convertit de réseau de neurones. Le second argument définit la pente dans la fonction d’activation.
net = NeuralTreeNet.create_from_tree(dec, 0.5)
net15 = NeuralTreeNet.create_from_tree(dec, 15)
from sklearn.metrics import accuracy_score
(roc_auc_score(clsy, dec.predict_proba(clsX)[:, 1]),
accuracy_score(clsy, dec.predict(clsX)))
(0.9141, 0.8733333333333333)
(roc_auc_score(clsy, net.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(net.predict(clsX)[:, -2:], axis=1)))
(0.8892, 0.7066666666666667)
(roc_auc_score(clsy, net15.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(net15.predict(clsX)[:, -2:], axis=1)))
(0.8876, 0.8666666666666667)
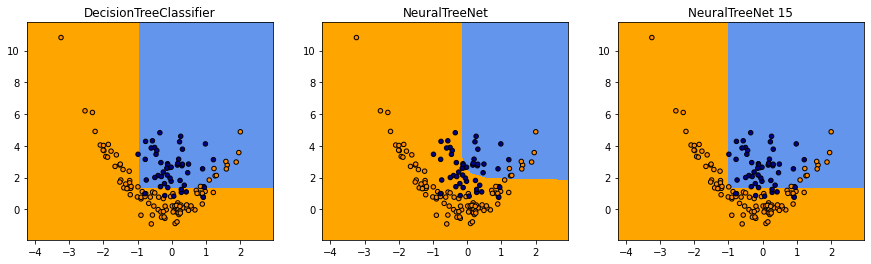
Le réseau de neurones est plus ou moins performant selon la pente dans la fonction d’activation.
fig, ax = plt.subplots(1, 3, figsize=(15, 4))
plot_grid(clsX, clsy, dec.predict, dec.__class__.__name__, ax=ax[0])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net.predict(x)[:, -2:], axis=1),
net.__class__.__name__, ax=ax[1])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net15.predict(x)[:, -2:], axis=1),
net15.__class__.__name__ + ' 15', ax=ax[2])
Et on apprend le réseau de neurones en partant de l’arbre de départ. On choisit celui qui a la pente d’activation la plus faible.
from mlstatpy.ml.neural_tree import label_class_to_softmax_output
clsY = label_class_to_softmax_output(clsy)
clsY[:3]
array([[1., 0.],
[1., 0.],
[1., 0.]])
net2 = net.copy()
net2.fit(clsX, clsY, verbose=True, max_iter=25, lr=3e-6)
0/25: loss: 746.7 lr=3e-06 max(coef): 1 l1=0/15 l2=0/9.8
1/25: loss: 728.8 lr=2.44e-07 max(coef): 1 l1=2.8e+02/15 l2=4.3e+03/9.6
2/25: loss: 726.5 lr=1.73e-07 max(coef): 1 l1=2.8e+02/15 l2=4.4e+03/9.6
3/25: loss: 726 lr=1.41e-07 max(coef): 1 l1=2.7e+02/15 l2=3.4e+03/9.6
4/25: loss: 725.2 lr=1.22e-07 max(coef): 1 l1=2.4e+02/15 l2=3.3e+03/9.6
5/25: loss: 724.9 lr=1.09e-07 max(coef): 1.1 l1=2.4e+02/15 l2=3.3e+03/9.6
6/25: loss: 724.4 lr=9.99e-08 max(coef): 1.1 l1=2.5e+02/15 l2=4.2e+03/9.6
7/25: loss: 724 lr=9.25e-08 max(coef): 1.1 l1=1.9e+02/15 l2=3.4e+03/9.6
8/25: loss: 723.7 lr=8.66e-08 max(coef): 1.1 l1=2.5e+02/15 l2=4.2e+03/9.6
9/25: loss: 723.4 lr=8.16e-08 max(coef): 1.1 l1=3.1e+02/15 l2=5.1e+03/9.6
10/25: loss: 723.2 lr=7.74e-08 max(coef): 1.1 l1=2.8e+02/15 l2=4.3e+03/9.6
11/25: loss: 722.9 lr=7.38e-08 max(coef): 1.1 l1=1.9e+02/15 l2=3.1e+03/9.6
12/25: loss: 722.7 lr=7.07e-08 max(coef): 1.1 l1=2.9e+02/15 l2=4.1e+03/9.6
13/25: loss: 722.5 lr=6.79e-08 max(coef): 1.1 l1=2.9e+02/15 l2=4e+03/9.6
14/25: loss: 722.3 lr=6.54e-08 max(coef): 1.1 l1=2.9e+02/15 l2=4.6e+03/9.6
15/25: loss: 722.1 lr=6.32e-08 max(coef): 1.1 l1=3e+02/15 l2=3.9e+03/9.6
16/25: loss: 722 lr=6.12e-08 max(coef): 1.1 l1=2.9e+02/15 l2=4e+03/9.6
17/25: loss: 721.8 lr=5.94e-08 max(coef): 1.1 l1=3e+02/15 l2=3.9e+03/9.6
18/25: loss: 721.7 lr=5.77e-08 max(coef): 1.1 l1=2e+02/15 l2=2.9e+03/9.6
19/25: loss: 721.6 lr=5.62e-08 max(coef): 1.1 l1=3e+02/15 l2=4.5e+03/9.6
20/25: loss: 721.5 lr=5.48e-08 max(coef): 1.1 l1=2.8e+02/15 l2=4.1e+03/9.6
21/25: loss: 721.3 lr=5.34e-08 max(coef): 1.1 l1=3.1e+02/15 l2=5.1e+03/9.6
22/25: loss: 721.2 lr=5.22e-08 max(coef): 1.1 l1=3e+02/15 l2=4.9e+03/9.6
23/25: loss: 721.1 lr=5.11e-08 max(coef): 1.1 l1=2.5e+02/15 l2=4.6e+03/9.6
24/25: loss: 721 lr=5e-08 max(coef): 1.1 l1=2.5e+02/15 l2=3.7e+03/9.6
25/25: loss: 721 lr=4.9e-08 max(coef): 1.1 l1=2e+02/15 l2=3.6e+03/9.6
NeuralTreeNet(2)
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net.predict(x)[:, -2:], axis=1),
"Avant apprentissage", ax=ax[0])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net2.predict(x)[:, -2:], axis=1),
"Après apprentissage", ax=ax[1])
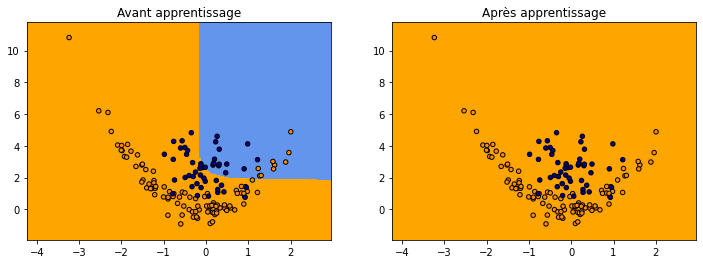
Ca ne marche pas ou pas très bien. Il faudrait vérifier que la configuration actuelle ne se trouve pas dans un minimum local auquel cas l’apprentissage par gradient ne donnera quasiment rien.
(roc_auc_score(clsy, net2.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(net2.predict(clsX)[:, -2:], axis=1)))
(0.8818, 0.6666666666666666)
net2.predict(clsX)[-5:, -2:]
array([[0.66986362, 0.33013638],
[0.716717 , 0.283283 ],
[0.58034967, 0.41965033],
[0.74722945, 0.25277055],
[0.55707209, 0.44292791]])
net.predict(clsX)[-5:, -2:]
array([[0.59071361, 0.40928639],
[0.63784777, 0.36215223],
[0.49777366, 0.50222634],
[0.66626214, 0.33373786],
[0.47202372, 0.52797628]])
On peut essayer de repartir à zéro. Des fois ça peut marcher mais il faudrait beaucoup plus d’essai.
net3 = net.copy()
dim = net3.training_weights.shape
net3.update_training_weights(numpy.random.randn(dim[0]))
net3.fit(clsX, clsY, verbose=True, max_iter=25, lr=3e-6)
0/25: loss: 1594 lr=3e-06 max(coef): 2.5 l1=0/27 l2=0/37
1/25: loss: 1514 lr=2.44e-07 max(coef): 2.5 l1=7.8e+02/25 l2=8.1e+04/34
2/25: loss: 1509 lr=1.73e-07 max(coef): 2.5 l1=3.2e+02/25 l2=6.9e+03/34
3/25: loss: 1497 lr=1.41e-07 max(coef): 2.5 l1=6.3e+02/25 l2=2.8e+04/33
4/25: loss: 1498 lr=1.22e-07 max(coef): 2.5 l1=6.7e+02/25 l2=3.3e+04/33
5/25: loss: 1509 lr=1.09e-07 max(coef): 2.5 l1=5.6e+02/25 l2=3.7e+04/33
6/25: loss: 1521 lr=9.99e-08 max(coef): 2.5 l1=8.8e+02/25 l2=1.2e+05/33
7/25: loss: 1526 lr=9.25e-08 max(coef): 2.5 l1=1.1e+03/25 l2=2.7e+05/33
8/25: loss: 1529 lr=8.66e-08 max(coef): 2.5 l1=9e+02/25 l2=1.2e+05/33
9/25: loss: 1534 lr=8.16e-08 max(coef): 2.5 l1=1.6e+03/25 l2=7.2e+05/33
10/25: loss: 1540 lr=7.74e-08 max(coef): 2.5 l1=6.3e+02/25 l2=4.7e+04/33
11/25: loss: 1539 lr=7.38e-08 max(coef): 2.5 l1=1.1e+03/25 l2=2.1e+05/33
12/25: loss: 1538 lr=7.07e-08 max(coef): 2.5 l1=7.8e+02/25 l2=7.7e+04/33
13/25: loss: 1542 lr=6.79e-08 max(coef): 2.5 l1=6.8e+02/25 l2=5.5e+04/33
14/25: loss: 1547 lr=6.54e-08 max(coef): 2.5 l1=5.6e+02/25 l2=3.7e+04/33
15/25: loss: 1553 lr=6.32e-08 max(coef): 2.5 l1=5.4e+02/25 l2=3.5e+04/33
16/25: loss: 1559 lr=6.12e-08 max(coef): 2.5 l1=6.4e+02/25 l2=4.4e+04/33
17/25: loss: 1572 lr=5.94e-08 max(coef): 2.5 l1=1.1e+03/25 l2=2.6e+05/33
18/25: loss: 1583 lr=5.77e-08 max(coef): 2.5 l1=5.2e+02/25 l2=3.4e+04/33
19/25: loss: 1588 lr=5.62e-08 max(coef): 2.5 l1=7e+02/25 l2=6.2e+04/33
20/25: loss: 1599 lr=5.48e-08 max(coef): 2.5 l1=1.3e+03/25 l2=3.6e+05/33
21/25: loss: 1608 lr=5.34e-08 max(coef): 2.5 l1=6e+02/25 l2=2.4e+04/33
22/25: loss: 1619 lr=5.22e-08 max(coef): 2.5 l1=1.3e+03/25 l2=4.8e+05/33
23/25: loss: 1623 lr=5.11e-08 max(coef): 2.5 l1=7e+02/25 l2=4.3e+04/33
24/25: loss: 1629 lr=5e-08 max(coef): 2.5 l1=4.9e+02/25 l2=1.8e+04/33
25/25: loss: 1635 lr=4.9e-08 max(coef): 2.5 l1=3.1e+02/25 l2=6.3e+03/33
NeuralTreeNet(2)
(roc_auc_score(clsy, net3.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(net3.predict(clsX)[:, -2:], axis=1)))
(0.3596, 0.38)
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net.predict(x)[:, -2:], axis=1),
"Avant apprentissage", ax=ax[0])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(net3.predict(x)[:, -2:], axis=1),
"Après apprentissage", ax=ax[1])
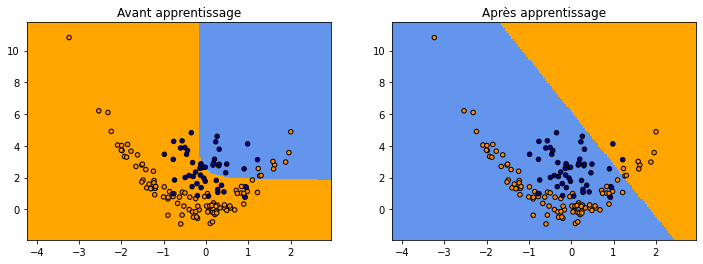
Autre architecture#
Cette fois-ci, on réduit le nombre de neurones. Au lieu d’avoir deux neurones par noeud du graphe, on assemble tous les neurones en deux : un pour les entrées, un autre pour le calcul des sorties. Les deux représentations ne sont pas implémentées de façon rigoureusement identique dans le module mlstatpy. Le code précise les différences.
netc = NeuralTreeNet.create_from_tree(dec, 1, arch='compact')
RenderJsDot(netc.to_dot())
(roc_auc_score(clsy, netc.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(netc.predict(clsX)[:, -2:], axis=1)))
(0.8937999999999999, 0.3333333333333333)
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_grid(clsX, clsy,
lambda x: numpy.argmax(dec.predict_proba(x), axis=1),
"Avant conversion", ax=ax[0])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(netc.predict(x)[:, -2:], axis=1),
"Après comversion", ax=ax[1])
On réapprend.
netc4 = netc.copy()
netc4.fit(clsX, clsY, verbose=True, max_iter=25, lr=1e-6)
0/25: loss: 1278 lr=1e-06 max(coef): 1.5 l1=0/27 l2=0/28
1/25: loss: 899 lr=8.14e-08 max(coef): 1.5 l1=9.7e+02/27 l2=5.5e+04/27
2/25: loss: 852.3 lr=5.76e-08 max(coef): 1.5 l1=1.1e+02/27 l2=1.3e+03/27
3/25: loss: 833.7 lr=4.71e-08 max(coef): 1.5 l1=1.1e+02/27 l2=1.5e+03/27
4/25: loss: 821 lr=4.08e-08 max(coef): 1.5 l1=5.4e+02/27 l2=1.6e+04/27
5/25: loss: 808.7 lr=3.65e-08 max(coef): 1.5 l1=1.8e+02/27 l2=2.7e+03/27
6/25: loss: 799 lr=3.33e-08 max(coef): 1.5 l1=1.1e+02/27 l2=1.4e+03/27
7/25: loss: 791.2 lr=3.08e-08 max(coef): 1.5 l1=7.1e+02/27 l2=2.8e+04/27
8/25: loss: 784.5 lr=2.89e-08 max(coef): 1.5 l1=4.8e+02/27 l2=1.3e+04/27
9/25: loss: 777.2 lr=2.72e-08 max(coef): 1.5 l1=1.7e+03/27 l2=3.8e+05/27
10/25: loss: 771.3 lr=2.58e-08 max(coef): 1.5 l1=5.6e+02/27 l2=1.7e+04/27
11/25: loss: 765 lr=2.46e-08 max(coef): 1.5 l1=1.4e+03/27 l2=2.1e+05/27
12/25: loss: 760.1 lr=2.36e-08 max(coef): 1.5 l1=1.7e+03/27 l2=3.8e+05/27
13/25: loss: 755.4 lr=2.26e-08 max(coef): 1.5 l1=6e+02/27 l2=1.7e+04/27
14/25: loss: 750.5 lr=2.18e-08 max(coef): 1.5 l1=5.1e+02/27 l2=1.5e+04/27
15/25: loss: 746.6 lr=2.11e-08 max(coef): 1.5 l1=2.6e+02/27 l2=5.5e+03/27
16/25: loss: 742.6 lr=2.04e-08 max(coef): 1.5 l1=5.2e+02/27 l2=1.2e+04/27
17/25: loss: 739.1 lr=1.98e-08 max(coef): 1.5 l1=5.2e+02/27 l2=1.6e+04/27
18/25: loss: 735.4 lr=1.92e-08 max(coef): 1.5 l1=6.2e+02/27 l2=1.9e+04/27
19/25: loss: 731.9 lr=1.87e-08 max(coef): 1.5 l1=5.1e+02/27 l2=1.7e+04/27
20/25: loss: 728.5 lr=1.83e-08 max(coef): 1.5 l1=1.2e+02/27 l2=1.6e+03/27
21/25: loss: 725 lr=1.78e-08 max(coef): 1.5 l1=4.2e+02/27 l2=1.1e+04/27
22/25: loss: 721.4 lr=1.74e-08 max(coef): 1.5 l1=5.1e+02/27 l2=1.1e+04/27
23/25: loss: 718.7 lr=1.7e-08 max(coef): 1.5 l1=1.1e+03/27 l2=1.5e+05/27
24/25: loss: 715.8 lr=1.67e-08 max(coef): 1.5 l1=1.5e+02/27 l2=2.8e+03/27
25/25: loss: 713 lr=1.63e-08 max(coef): 1.5 l1=1.3e+02/27 l2=1.7e+03/27
NeuralTreeNet(2)
(roc_auc_score(clsy, netc4.predict(clsX)[:, -1]),
accuracy_score(clsy, numpy.argmax(netc4.predict(clsX)[:, -2:], axis=1)))
(0.8603999999999999, 0.8466666666666667)
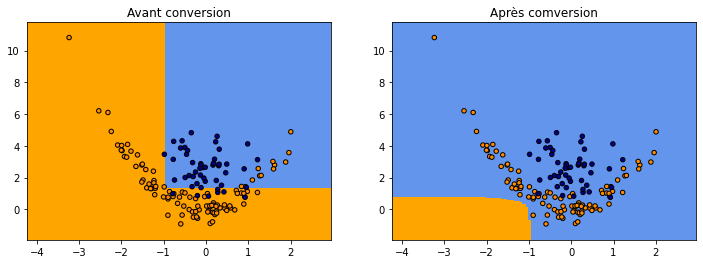
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
plot_grid(clsX, clsy,
lambda x: numpy.argmax(netc.predict(x)[:, -2:], axis=1),
"Avant apprentissage", ax=ax[0])
plot_grid(clsX, clsy,
lambda x: numpy.argmax(netc4.predict(x)[:, -2:], axis=1),
"Après apprentissage", ax=ax[1])
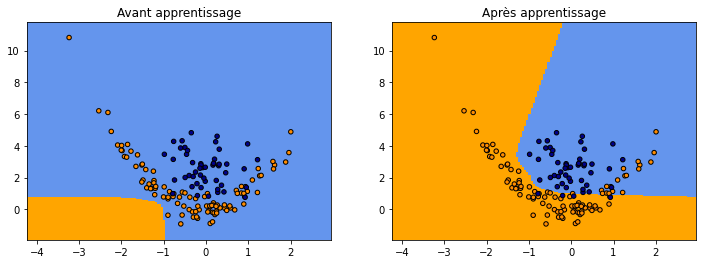
C’est mieux…