Un exemple simple de régression linéaire#
Links: notebook, html, PDF, python, slides, GitHub
Un jeu de données, une régression linéaire et quelques trucs bizarres. Le jeu de données est Wine Data Set.
%matplotlib inline
from jyquickhelper import add_notebook_menu
add_notebook_menu()
Récupération des données#
Le premier code récupère les données depuis Internet.
import pandas
df = pandas.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data",
header=None)
df.head(n=1)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 14.23 | 1.71 | 2.43 | 15.6 | 127 | 2.8 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065 |
df.shape
(178, 14)
On récupère le nom des colonnes.
s = "index Alcohol Malicacid Ash Alcalinityofash Magnesium Totalphenols Flavanoids"
s += " Nonflavanoidphenols Proanthocyanins Colorintensity Hue"
s += " OD280/OD315 Proline"
df.columns = s.split()
df.head().T
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| index | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Alcohol | 14.23 | 13.20 | 13.16 | 14.37 | 13.24 |
| Malicacid | 1.71 | 1.78 | 2.36 | 1.95 | 2.59 |
| Ash | 2.43 | 2.14 | 2.67 | 2.50 | 2.87 |
| Alcalinityofash | 15.60 | 11.20 | 18.60 | 16.80 | 21.00 |
| Magnesium | 127.00 | 100.00 | 101.00 | 113.00 | 118.00 |
| Totalphenols | 2.80 | 2.65 | 2.80 | 3.85 | 2.80 |
| Flavanoids | 3.06 | 2.76 | 3.24 | 3.49 | 2.69 |
| Nonflavanoidphenols | 0.28 | 0.26 | 0.30 | 0.24 | 0.39 |
| Proanthocyanins | 2.29 | 1.28 | 2.81 | 2.18 | 1.82 |
| Colorintensity | 5.64 | 4.38 | 5.68 | 7.80 | 4.32 |
| Hue | 1.04 | 1.05 | 1.03 | 0.86 | 1.04 |
| OD280/OD315 | 3.92 | 3.40 | 3.17 | 3.45 | 2.93 |
| Proline | 1065.00 | 1050.00 | 1185.00 | 1480.00 | 735.00 |
df.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| index | 178.0 | 1.938202 | 0.775035 | 1.00 | 1.0000 | 2.000 | 3.0000 | 3.00 |
| Alcohol | 178.0 | 13.000618 | 0.811827 | 11.03 | 12.3625 | 13.050 | 13.6775 | 14.83 |
| Malicacid | 178.0 | 2.336348 | 1.117146 | 0.74 | 1.6025 | 1.865 | 3.0825 | 5.80 |
| Ash | 178.0 | 2.366517 | 0.274344 | 1.36 | 2.2100 | 2.360 | 2.5575 | 3.23 |
| Alcalinityofash | 178.0 | 19.494944 | 3.339564 | 10.60 | 17.2000 | 19.500 | 21.5000 | 30.00 |
| Magnesium | 178.0 | 99.741573 | 14.282484 | 70.00 | 88.0000 | 98.000 | 107.0000 | 162.00 |
| Totalphenols | 178.0 | 2.295112 | 0.625851 | 0.98 | 1.7425 | 2.355 | 2.8000 | 3.88 |
| Flavanoids | 178.0 | 2.029270 | 0.998859 | 0.34 | 1.2050 | 2.135 | 2.8750 | 5.08 |
| Nonflavanoidphenols | 178.0 | 0.361854 | 0.124453 | 0.13 | 0.2700 | 0.340 | 0.4375 | 0.66 |
| Proanthocyanins | 178.0 | 1.590899 | 0.572359 | 0.41 | 1.2500 | 1.555 | 1.9500 | 3.58 |
| Colorintensity | 178.0 | 5.058090 | 2.318286 | 1.28 | 3.2200 | 4.690 | 6.2000 | 13.00 |
| Hue | 178.0 | 0.957449 | 0.228572 | 0.48 | 0.7825 | 0.965 | 1.1200 | 1.71 |
| OD280/OD315 | 178.0 | 2.611685 | 0.709990 | 1.27 | 1.9375 | 2.780 | 3.1700 | 4.00 |
| Proline | 178.0 | 746.893258 | 314.907474 | 278.00 | 500.5000 | 673.500 | 985.0000 | 1680.00 |
Corrélations#
On utilise le module seaborn pour regarder visuellement les corrélations avec le graphe Scatterplot Matrix.
import seaborn as sns
sns.set(style="ticks")
sns.pairplot(df);
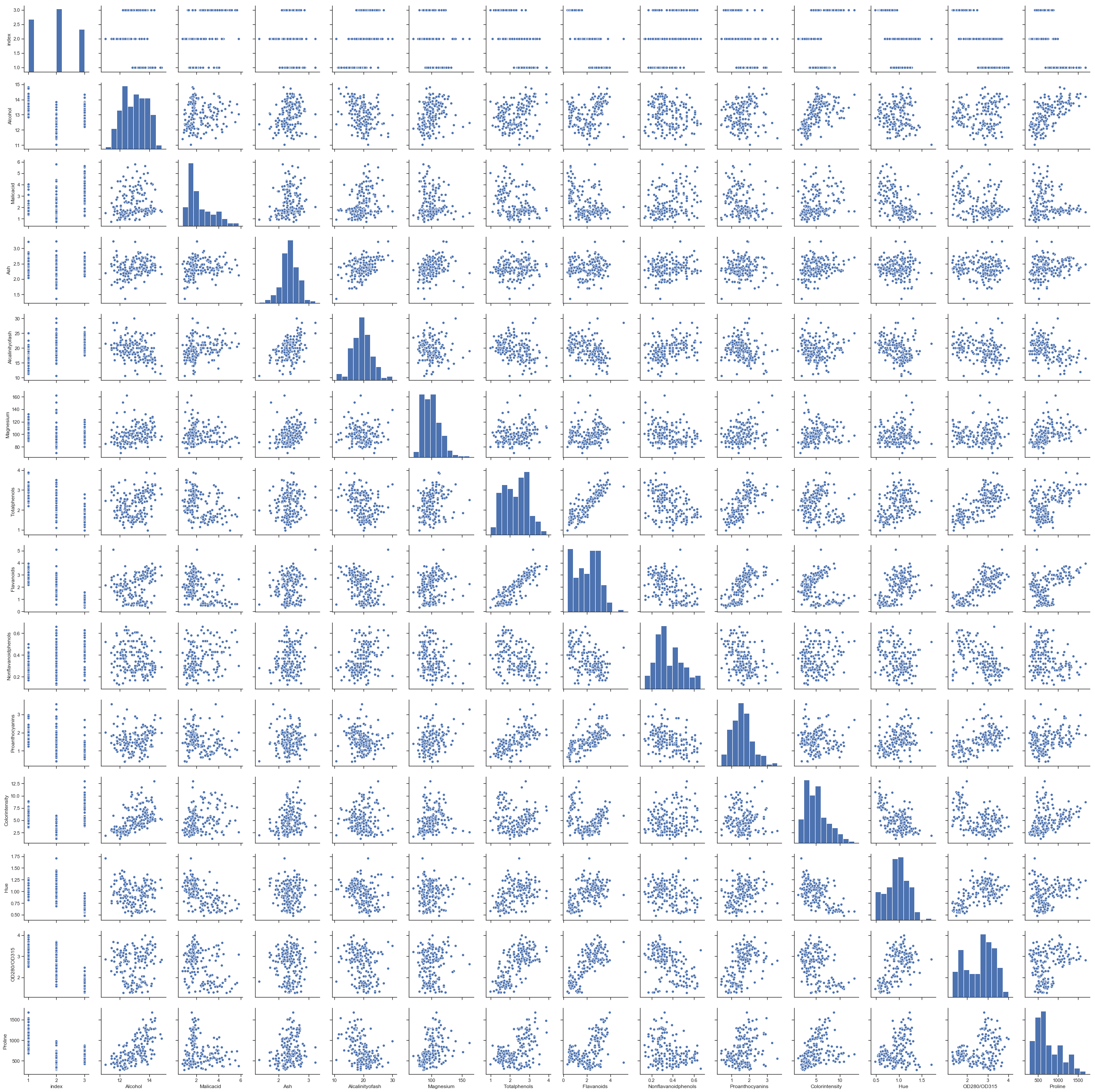
Il y a quelques corrélations.
Première régression#
On essaye de prédire le taux d’alcool du vin à partir des autres variables.
X = df.drop(["Alcohol", "index"], axis=1)
y = df["Alcohol"]
On divise entre train et test pour éviter l’overfitting.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train, y_train)
LinearRegression(copy_X=True, fit_intercept=True, n_jobs=None,
normalize=False)
model.coef_
array([ 1.63363895e-01, 3.94203469e-01, -6.00949512e-02, -1.19824047e-04,
1.58697845e-01, -9.56728506e-02, -3.06298297e-01, 5.07946728e-02,
1.38811432e-01, 1.59459210e-02, 1.64240691e-01, 8.15269173e-04])
from sklearn.metrics import r2_score
y_pred = model.predict(X_test)
r2_score(y_test, y_pred)
0.4649938412573541
y_predt = model.predict(X_train)
r2_score(y_train, y_predt)
0.6136769355983627
Même chose avec un arbre de décision.
from sklearn.tree import DecisionTreeRegressor
dt = DecisionTreeRegressor(min_samples_leaf=10)
dt.fit(X_train, y_train)
DecisionTreeRegressor(criterion='mse', max_depth=None, max_features=None,
max_leaf_nodes=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=10,
min_samples_split=2, min_weight_fraction_leaf=0.0,
presort=False, random_state=None, splitter='best')
y_pred = dt.predict(X_test)
r2_score(y_test, y_pred)
0.6694565403724826
y_predt = dt.predict(X_train)
r2_score(y_train, y_predt)
0.7172799827093439
X_train.shape
(133, 12)
On vérifie avec une validation croisée.
from sklearn.model_selection import cross_val_score
cross_val_score(model, X, y, cv=5)
array([-0.51847929, 0.40456596, -0.73716381, -0.46995099, -0.47213708])
Ca oscille trop pour que cela soit vraisemblable. C’est probablement dû à l’ordre dans les données.
try:
from sklearn.utils import shuffle
df2 = shuffle(df).reset_index()
except Exception:
import random
index = list(df.index)
df2 = df.iloc[index].reset_index()
X2 = df2.drop("Alcohol", axis=1)
y2 = df2.Alcohol
cross_val_score(model, X2, y2, cv=5)
array([0.40197927, 0.55886103, 0.57939265, 0.49028122, 0.62212365])
Beaucoup plus stable.
Feature importance#
dt.feature_importances_
array([0.03523663, 0. , 0.00776046, 0. , 0. ,
0. , 0.02238126, 0. , 0.6849213 , 0. ,
0.0124994 , 0.23720095])
ft = pandas.DataFrame(dict(name=X_train.columns, fi=dt.feature_importances_))
ft = ft.sort_values("fi", ascending=False)
ft
| name | fi | |
|---|---|---|
| 8 | Colorintensity | 0.684921 |
| 11 | Proline | 0.237201 |
| 0 | Malicacid | 0.035237 |
| 6 | Nonflavanoidphenols | 0.022381 |
| 10 | OD280/OD315 | 0.012499 |
| 2 | Alcalinityofash | 0.007760 |
| 1 | Ash | 0.000000 |
| 3 | Magnesium | 0.000000 |
| 4 | Totalphenols | 0.000000 |
| 5 | Flavanoids | 0.000000 |
| 7 | Proanthocyanins | 0.000000 |
| 9 | Hue | 0.000000 |
ft.set_index("name").plot(kind="bar");
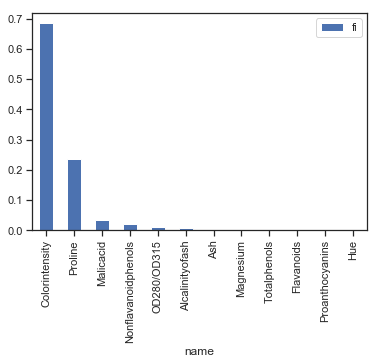
L’intensité de la couleur a l’air de jouer un grand rôle mais dans quel sens ?
ax = df[["Colorintensity", "Alcohol"]].plot(x="Colorintensity", y="Alcohol", kind="scatter")
ax.set_title("Lien entre le degré d'alcool\net l'intensité de la couleur");
'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.
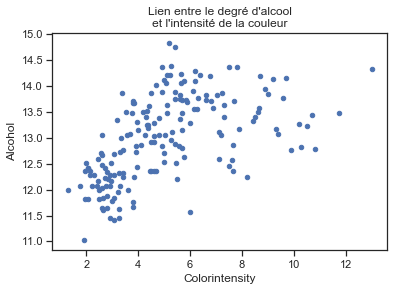
La relation n’est pas linéaire car le degré d’alcool montre rarement au-dessus de 14.5 mais elle paraît linéaire par morceaux.