Note
Click here to download the full example code
ONNX in a torch function#
The ONNX graph used in this example is not really interesting but it shows how to create a custom autograd function following torch API (Extending Pytorch). Methods forward and backward are implemented with ONNX and onnxruntime-training.
The ONNX Graph#
import pprint
import logging
import numpy
from pandas import DataFrame
import matplotlib.pyplot as plt
from skl2onnx.common.data_types import FloatTensorType
from skl2onnx.algebra.onnx_ops import (
OnnxSigmoid, OnnxMatMul, OnnxAdd)
from tqdm import tqdm
from mlprodict.onnx_tools.onnx_manipulations import onnx_rename_names
from mlprodict.plotting.plotting_onnx import plot_onnx
import torch
from deeponnxcustom.onnxtorch.torchort import TorchOrtFactory
def from_numpy(v, device=None, requires_grad=False):
"""
Convers a numpy array into a torch array and
sets *device* and *requires_grad*.
"""
v = torch.from_numpy(v)
if device is not None:
v = v.to(device)
v.requires_grad_(requires_grad)
return v
def create_onnx_graph(N, d_in=3, d_out=2, n_loops=1, opv=14):
"""
Returns a weird ONNX graph and its weights.
"""
var = [('X', FloatTensorType([N, d_in]))]
sum_node = None
weights_values = []
for i in range(n_loops):
cst = numpy.random.randn(d_in, 1).astype(numpy.float32) / (i + 1)
weights_values.append(cst)
mul = OnnxMatMul(var[0], cst, op_version=opv)
tanh = OnnxSigmoid(mul, op_version=opv)
if sum_node is None:
sum_node = tanh
else:
sum_node = OnnxAdd(sum_node, tanh, op_version=opv)
cst_mul = numpy.random.randn(1, d_out).astype(numpy.float32)
weights_values.append(cst_mul)
mul = OnnxMatMul(sum_node, cst_mul, op_version=opv)
cst_add = numpy.random.randn(1, d_out).astype(numpy.float32)
weights_values.append(cst_add)
final = OnnxAdd(mul, cst_add, op_version=opv, output_names=['Y'])
onx = final.to_onnx(
var, target_opset=opv, outputs=[('Y', FloatTensorType())])
weights_name = [i.name for i in onx.graph.initializer]
new_names = ['W%03d' % i for i in range(len(weights_name))]
onx = onnx_rename_names(onx, replace=dict(zip(weights_name, new_names)))
weights = list(zip(new_names, weights_values))
return onx, weights
Let’s see how it looks like.
N, d_in, d_out = 5, 3, 2
enable_logging = False
onx, weights = create_onnx_graph(N)
with open("plot_torch_ort.onnx", "wb") as f:
f.write(onx.SerializeToString())
print("weights to train:", [(w[0], w[1].shape) for w in weights])
ax = plot_onnx(onx)
ax.set_title("Model to train")
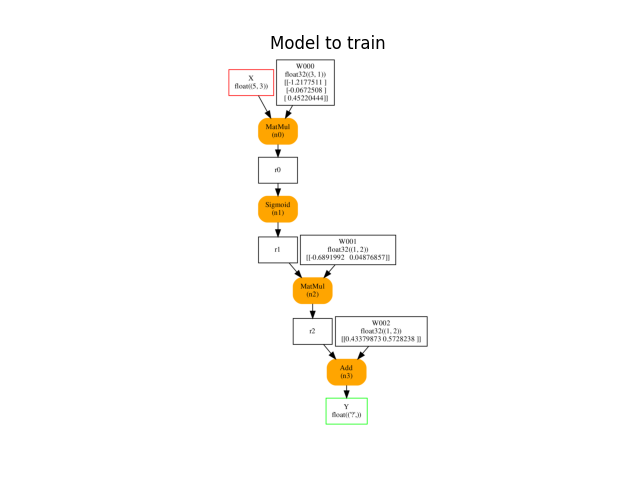
Out:
weights to train: [('W000', (3, 1)), ('W001', (1, 2)), ('W002', (1, 2))]
Text(0.5, 1.0, 'Model to train')
Wraps ONNX as a torch.autograd.Function#
Class TorchOrtFactory
uses onnxruntime-training to build the gradient with ONNX,
add calls it following this logic:
class CustomClass(torch.autograd.Function):
@staticmethod
def forward(ctx, *input):
ctx.save_for_backward(*input)
# inference with ONNX
return ...
@staticmethod
def backward(ctx, *grad_output):
input, = ctx.saved_tensors
# gradient with ONNX = inference with the gradient graph
return ...
The logic is hidden in TorchOrtFactory.create_class.
fact = TorchOrtFactory(onx, [w[0] for w in weights])
if enable_logging:
# Logging displays informations about the intermediate steps.
logger = logging.getLogger('deeponnxcustom')
logger.setLevel(logging.DEBUG)
logging.basicConfig(level=logging.DEBUG)
cls = fact.create_class(keep_models=True, enable_logging=enable_logging)
print(cls)
Out:
<class 'deeponnxcustom.onnxtorch.torchort.TorchOrtFunction_140651821009168'>
It produces the following inference graphs. The left one is the original one. The model on the left is the same except initializer are also inputs. If the input is missing, the initializer is considered as a default value.
fix, ax = plt.subplots(1, 2, figsize=(15, 10))
plot_onnx(onx, ax=ax[0])
plot_onnx(cls._optimized_pre_grad_model, ax=ax[1])
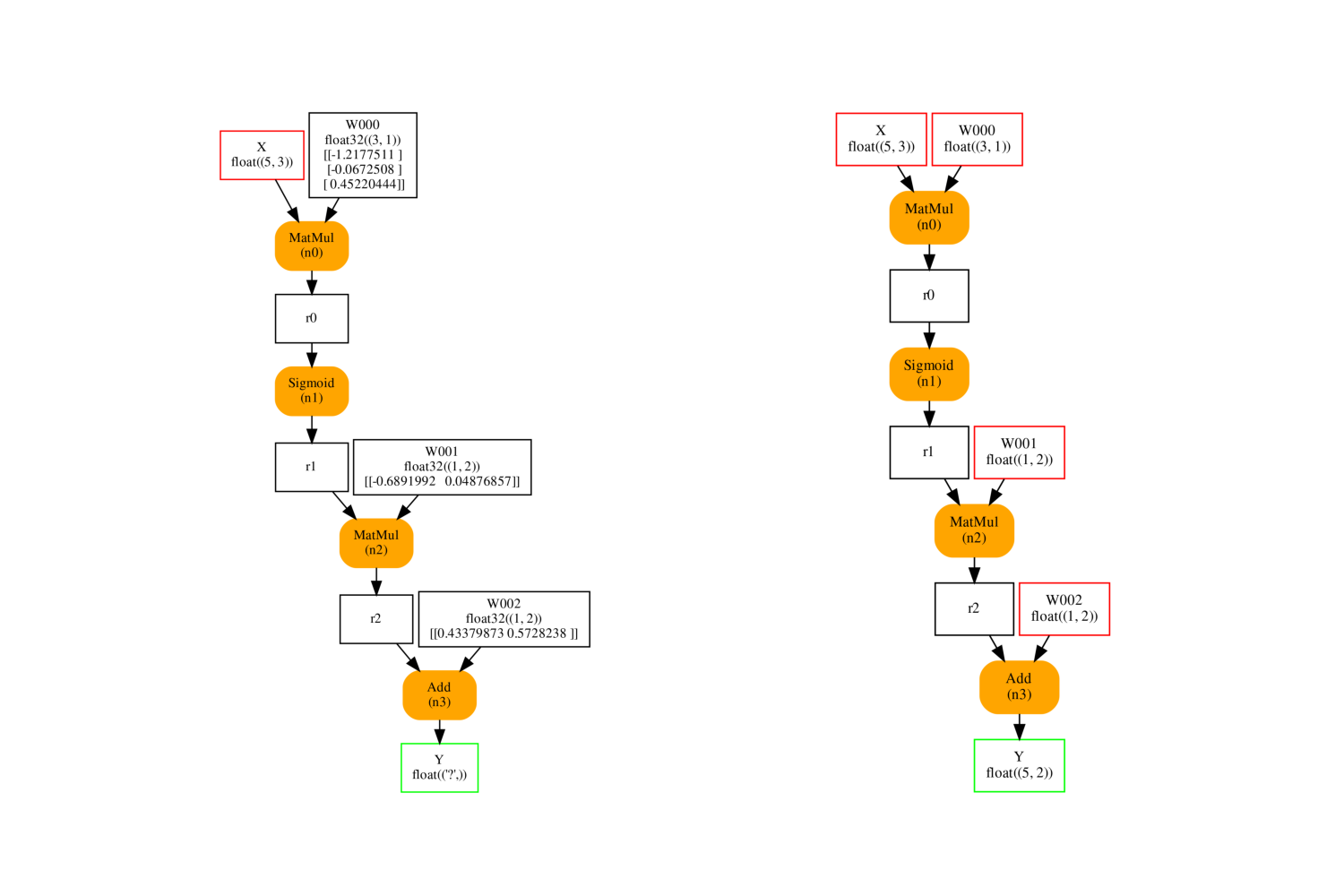
Out:
<AxesSubplot:>
And the gradient graph. It has the same inputs the previous graph on the right and has an output for every trained parameter.
fix, ax = plt.subplots(1, 1, figsize=(10, 10))
plot_onnx(cls._trained_onnx, ax=ax)
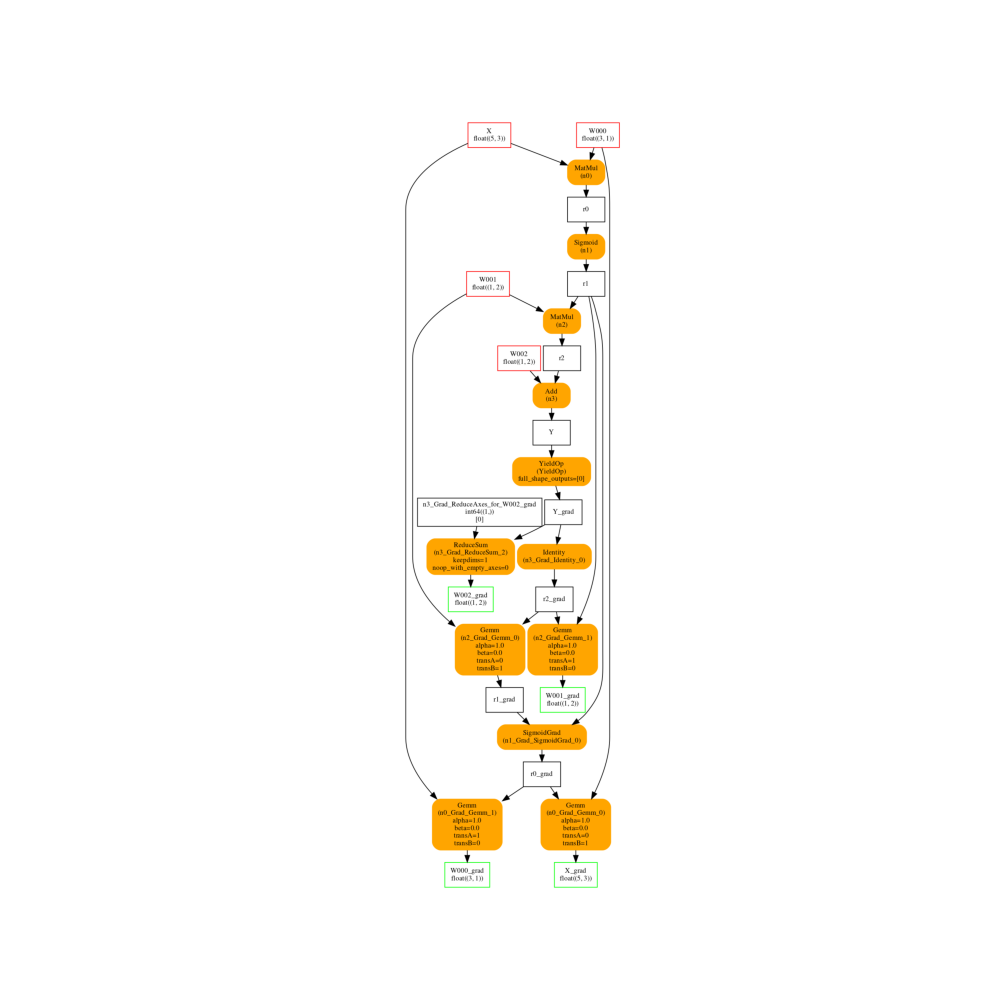
Out:
<AxesSubplot:>
Training#
We consider a simple example based on torch documentation (Learning Pytorch with Example or 110 - First percepton with pytorch).
def train_cls(cls, device, x, y, weights, n_iter=20, learning_rate=1e-2):
x = from_numpy(x, requires_grad=True, device=device)
y = from_numpy(y, requires_grad=True, device=device)
weights_tch = [(w[0], from_numpy(w[1], requires_grad=True, device=device))
for w in weights]
weights_values = [w[1] for w in weights_tch]
all_losses = []
for t in tqdm(range(n_iter)):
# forward - backward
y_pred = cls.apply(x, *weights_values)
loss = (y_pred - y).pow(2).sum()
loss.backward()
# update weights
with torch.no_grad():
for name, w in weights_tch:
w -= w.grad * learning_rate
w.grad.zero_()
all_losses.append((t, float(loss.cpu().detach().numpy())))
return all_losses, weights_tch
device_name = "cuda:0" if torch.cuda.is_available() else "cpu"
device = torch.device(device_name)
print("device:", device)
x = numpy.random.randn(N, d_in).astype(numpy.float32)
y = numpy.random.randn(N, d_out).astype(numpy.float32)
train_losses, final_weights = train_cls(cls, device, x, y, weights)
train_losses = numpy.array(train_losses)
pprint.pprint(final_weights)
Out:
device: cpu
0%| | 0/20 [00:00<?, ?it/s]
100%|##########| 20/20 [00:00<00:00, 464.60it/s]
[('W000',
tensor([[-1.2463],
[-0.0078],
[ 0.4170]], requires_grad=True)),
('W001', tensor([[-0.9162, -0.2614]], requires_grad=True)),
('W002', tensor([[0.2260, 0.2306]], requires_grad=True))]
The training loss is decreasing. The function seems to be learning something.
df = DataFrame(data=train_losses, columns=['iter', 'train_loss'])
df.plot(x="iter", y="train_loss", title="Training loss")
# plt.show()
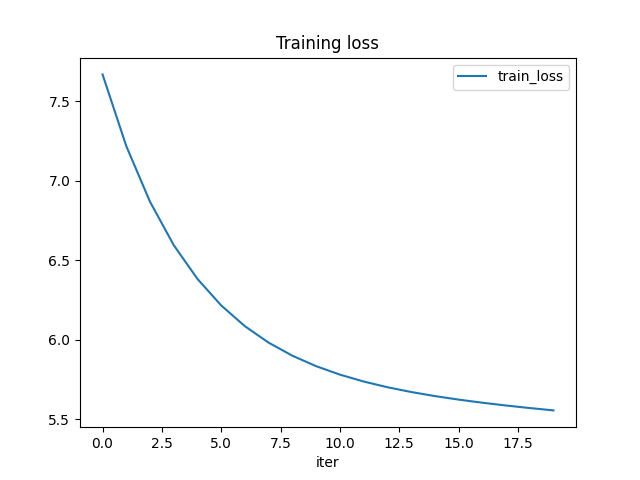
Out:
<AxesSubplot:title={'center':'Training loss'}, xlabel='iter'>
Total running time of the script: ( 0 minutes 6.746 seconds)